
Identify and select a region of the vector that you will replace with your insert. Set your backbone first using the pET-31b sequence. Open the “ Assembly Wizard” and choose “Gibson” as your assembly method. We’ll take a look at an example of Gibson cloning below where the JfyP gene is inserted into the pET 31b plasmid by designing sequence homology arms. Each DNA fragment possesses overlapping sequence homology that is used to direct the assembly reaction.
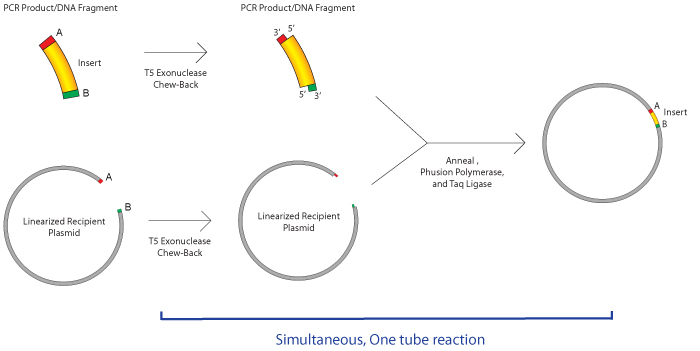
Daniel Gibson who developed this method to join multiple DNA fragments through a single isothermal reaction. Gibson assembly cloning is attributed to its creator Dr.
#BENCHLING VS SNAPGENE HOW TO#
Now find the area between your restriction sites that your JfyP insert will be cloned into and paste it in.įollow along with these step-by-step guides and videos to find out how to carry out Gibson and Golden Gate through Benchling’s Assembly Wizard. Identify where the ClaI and AlwNI restriction sites are in the pET-31b plasmid and ensure the JfyP gene insert possesses those restriction sites as well.Ĭopy the JfyP insert sequence and navigate back to your pET-31b sequence.Ĭreate a copy of your pET-31b sequence and rename it to include the JfyP gene you want to clone in. Practice Digest and Ligate Cloning - ManualĬan you recreate the pET-31b-JfyP construct by cloning manually and without the Benchling Assembly Wizard? Try to answer this question on your own and check the "Solution" at the bottom. When preparing for cloning, how could you check that the restriction enzymes have produced the desired DNA fragments? When you combine a DNA insert and vector, which of these enzyme would we use? A) DNA polymerase B) DNA ligase C) Restriction enzymeģ. In the above example, which component is the vector and which is the insert?Ģ. If you get a “green check” and the message “everything checks out” then you should name your new assembly and hit the green “ Assemble” button.Ĭhoose where to save your new molecule and Voila! You are done!ġ. Hold down “Ctrl+Shift” to click on the ClaI and AlwNI sites again. Now “right-click” to open a new menu and choose “Invert Selection” to highlight this area and choose “Set from Selection” to finalize your backbone.Ĭhoose the insert in your assembly and switch to the JfyP Insert sequence. Holding down “Ctrl+Shift” (or “Cmd + Shift” for Mac) and click on both the ClaI and AlwNI restriction sites to highlight the area between both restriction sites. Work on setting your backbone first and using the pET-31b sequence. Open the “ Assembly Wizard” and choose “Digest and Ligate” as your assembly method. Digest and Ligate Cloning - Assembly Wizardįollow the instructions below to use the Benchling Assembly Wizard to plan and design cloning for your DNA constructs. The fictional JfyP gene is inserted into the pET 31b plasmid using ClaI and AlwNI restriction sites.
We’ll look at an example of restriction cloning below. The cleavage can produce “sticky” or “blunt” ends of DNA that can be ligated to other DNA fragments that possess compatible ends. These enzymes cleave DNA sequences into fragments at or near specific recognition sites. Type II restriction enzymes are commonly used in restriction cloning workflows. We’ll first demonstrate how to do traditional restriction cloning (digest-and-ligate) using Benchling. Integrate plasmid maps into lab notebooks Plan and design molecular cloning strategiesĬompare different cloning methods including restriction, Gibson and Golden Gate cloning

See previous worksheets on Benchling that introduce PCR and restriction enzyme digests. A basic understanding of the concept of DNA cloning and familiarity with related concepts such as PCR, restriction enzymes, and DNA synthesis.


 0 kommentar(er)
0 kommentar(er)
